Step-1: Upload a VCF file:
Large files take longer to upload. However, you can also upload compressed versions of VCF files. Currently, VCF files compressed or archived into ‘VCF.gz’ and ‘VCF.tar.gz’ formats are supported.
Step-2: Select a Motif:
TraPS-VarI analyses VCF file to annotate all cell membrane proteins in the human genome. While annotating single pass transmembrane proteins, TraPS-VarI identifies membrane proximal tyrosine-based sequence motifs namely STAT3 docking site (YxxQ) in the juxtamembrane segment (Ulaganathan et al, Nature 2015), immunoreceptor tyrosine-based inhibition motif (S/I/V/LxYxxI/V/L) (Daëron et al, Immunity 1995), and immunoreceptor tyrosine-based activation motif (YxxI/Lx(6-30)YxxI/L) (Reth, Nature 1989). To identify genetic variations impacting any of these motifs, select your choice using the drop-down menu.
Step-3: Select the Assembly:
The human reference genome build used for generating the VCF file.
Step-4 & Step-5: Submitting the Task and Monitoring the Progress:
TraPS-VarI analysis of VCF file is a time-consuming task. To obtain notification by email, you can provide your email. Alternatively, enter a dummy email and check the status periodically by providing the Job ID.
Step-6: Navigating through the Results:
Here for illustration, the exome sequencing data of the acute lymphoblastic leukemia (ALL) cell line MOLT-4 (Reinhold et al, PLOS One 2014) was reanalyzed using the TraPS-VarI Web Application.
ITIM motif selected to identify the impact of germline variations on ITIM motifs and immunoreceptor protein domains.
(a) - ITIM Motif Analysis:
ITIM motif results show deletion of ITIM motif in SIT1, a T cell surface molecule that negatively regulates TCR (T-cell antigen receptor)-mediated signaling in T-cells. Involved in positive selection of T-cells. 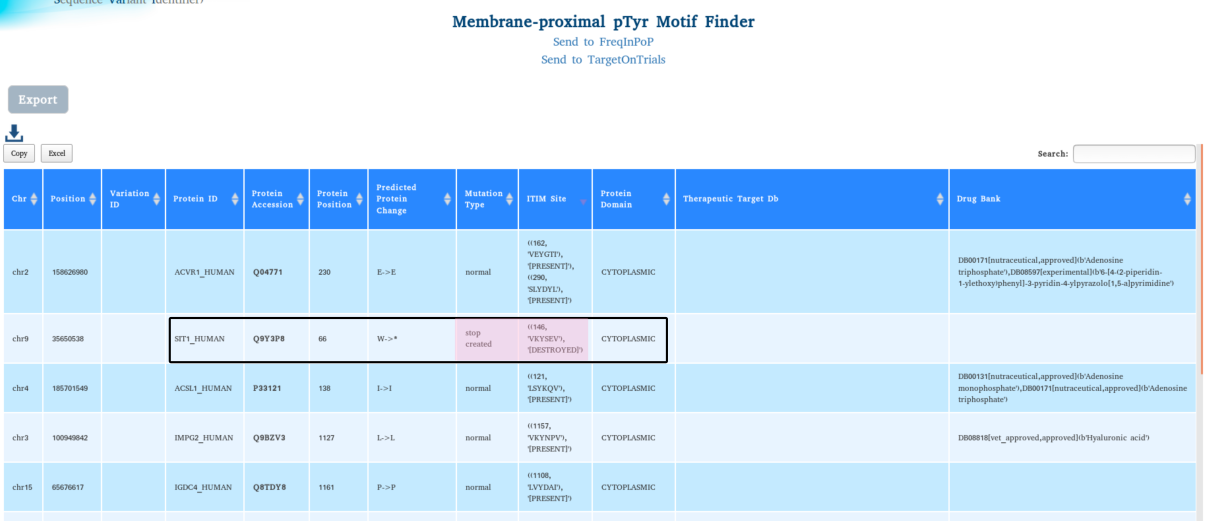
(b) - Mutation Mapping to Protein Domains:
MOLT-4 is an acute lymphoblastic leukemia (ALL) T lymphoblast cell line that was isolated from a 19 year male. The patient had a genetic variation in CTLA4 at the “SIGNAL” seqeunce p.T17A. 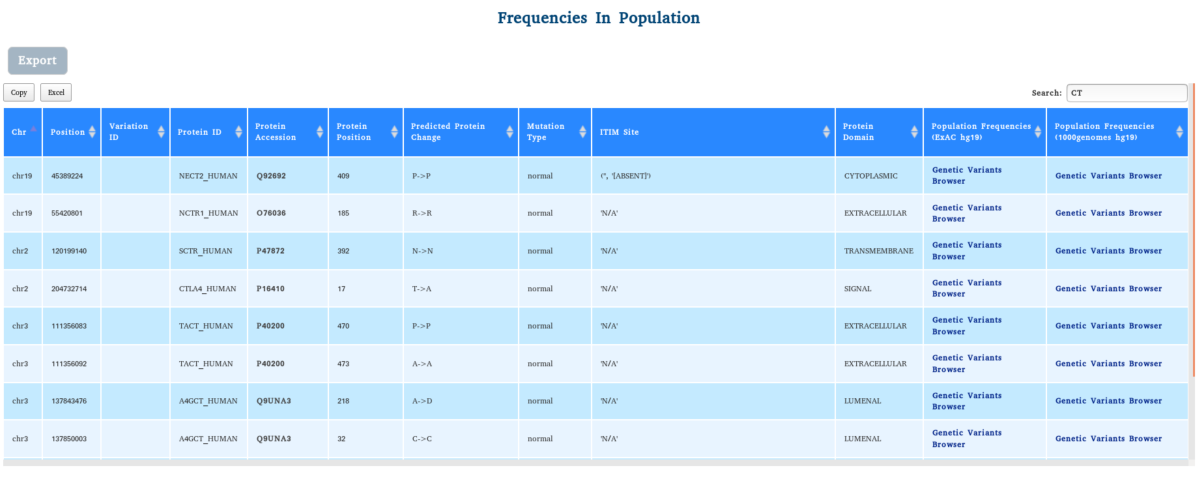
(c) - Allele Frequencies of Germline Variants:
For a global display of the information about the frequencies of membrane protein variants across the globe, the result table is directly linked to “Geography of Genetic Variants Browser (GGV)” . The GGV is developed by Joe Marcus and John Novembre with Jeff Tharsen and Alex Mueller, Department of Human Genetics, University of Chicago (https://popgen.uchicago.edu/ggv/)
The membrane protein variant CTLA4 p.T17A found in the ALL cell line MOLT-4 is a common variant widely distributed in the general population.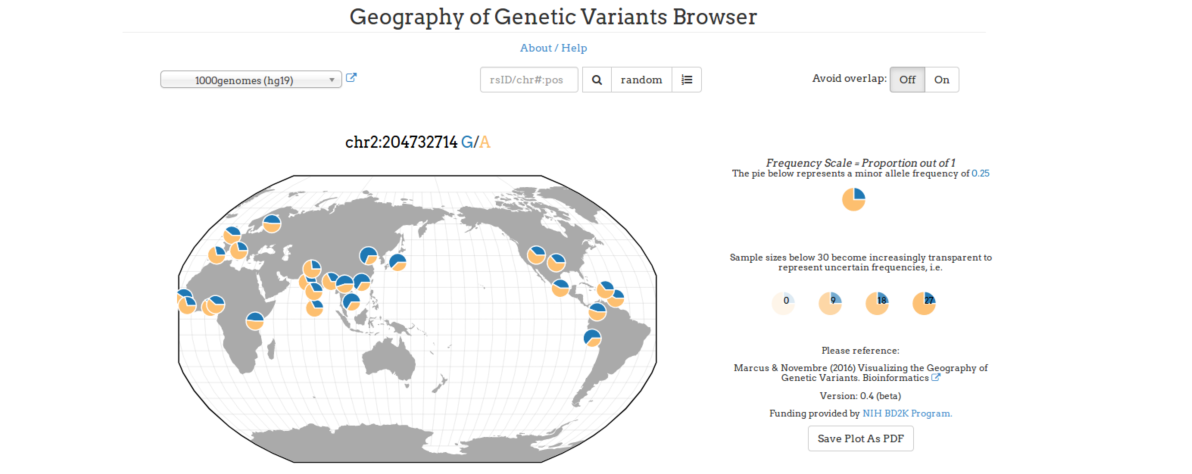
(d) - Clinical Trials for the Selected Membrane Protein Variant:
Ongoing Clinical Trial results to watch for potential effect of genetic variation on the outcomes of the pre-clinical and post-clinical studies. Since the germline mutation substitutes “Threonine” for “Alanine” at the position 17 in the ‘SIGNAL’ sequence of the membrane protein CTLA4, a potential impact on the surface expression and antibody binding can be expected. This information may be relevant for clinical trial studies.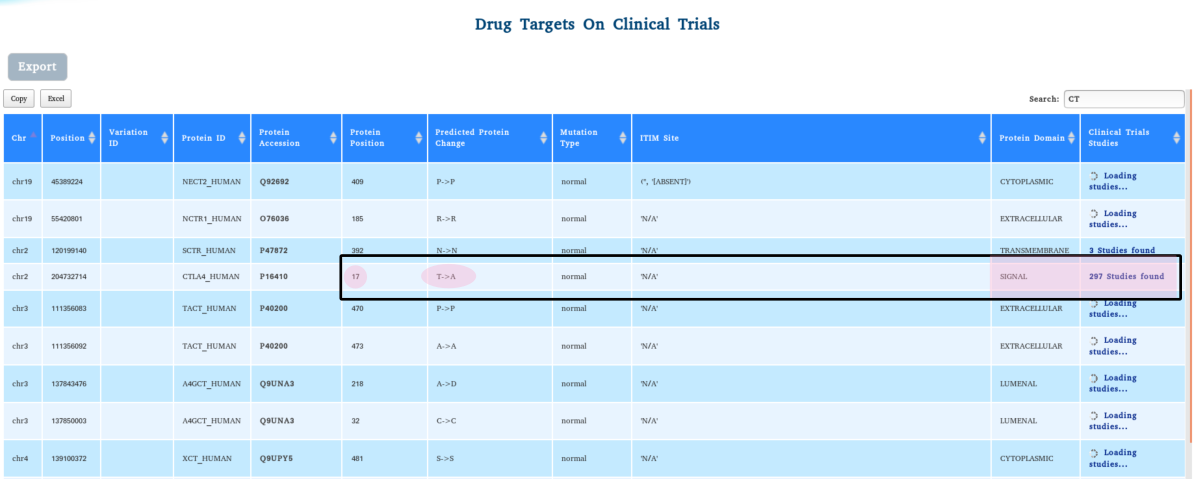
--
If you face any issues, please write to: help{at}traps-vari.org
Comments
Comments are closed.